The profiles are compared with reference samples for species identification. This procedure has been widely used in seafood authentication because it is less costly, simpler, and more suitable for routine laboratory analysis than techniques such as FINS or DNA barcoding, both of which are based on DNA sequencing analysis. PCR-RFLP is a relatively rapid, reproducible, and robust laboratory technique that does not require expensive equipment, and it is approved in many countries for the determination of seafood species. PCR-RFLP is therefore well suited for fish species detection, particularly for use closer to the origin of the sample.
Researchers at Campden BRI in England have developed a PCR-RFLP method that replaces the gel electrophoresis step with microfluidic lab-on-a-chip technology, utilizing CE to analyze DNA fragments.2-3 Lab-on-a-chip CE increases the ease of use, sensitivity, speed, and reliability of PCR-RFLP compared to gel-based methods.1 The chips are single-use units that contain etched capillaries attached directly to sample loading wells (see Figure 1, above).
The electrophoretic analysis and interpretation of results are completely automated, requiring only the click of a mouse after the samples are loaded onto the chip. The superior resolution of lab-on-a-chip technology enables the detection of DNA fragments that may be too small for visualization using gel electrophoresis. While the visual inspection of agarose gels and comparison to validated fish species patterns is tedious and error-prone, the lab-on-a-chip system automatically analyzes the pattern, compares it to a database of validated fish species patterns, and generates a species match. The database is expandable to thousands of species, assuring that this testing platform will be able to adapt to future needs.
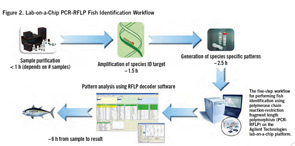
How It Works
The lab-on-a-chip analysis starts with the extraction of DNA, which requires only 40 mg to one gm of tissue and is done using a solid phase extraction spin column (see Figure 2, p. 22). The second step is the amplification of the target DNA sequence by PCR, involving a 464 bp segment of the cytochrome b gene (cytb), a gene that is found in all vertebrate fish, with a sequence that is known in a large number of fish species. The amplified DNA fragment is then digested with three restriction enzymes (DdeI, HaeIII, NiaIII) to generate fragment size patterns specific to the fish species. A kit that provides all the required reagents for the amplification and restriction digests comes with the lab-on-a-chip system.
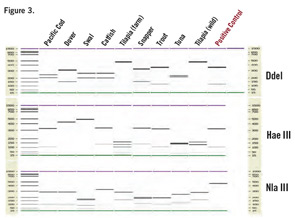
The fourth step involves loading the digested sample onto the chip and performing the automated electrophoretic separation of the restriction fragment. The final step is the automated analysis of the restriction fragment patterns by the onboard software that matches the patterns to a database of patterns from known species and identifies the most likely species (see Figure 3, below). The result is a fast and reliable species identification, even in samples from a mixture of fish species, with the ability to detect as little as 5% of a second species in the same sample.
Several laboratories have already adopted the lab-on-a-chip PCR-RFLP platform for fish species identification. For example, Eurofins Scientific, an international group of laboratories that provides testing and support services to the food industry, has been using this technique for some time at their Wolverhampton, England, facility. The company has found the Agilent platform very useful, in part because the fragment-matching software was extremely helpful due to the large database of species available.
ACCESS THE FULL VERSION OF THIS ARTICLE
To view this article and gain unlimited access to premium content on the FQ&S website, register for your FREE account. Build your profile and create a personalized experience today! Sign up is easy!
GET STARTED
Already have an account? LOGIN
